Quick Start#
nmma provides a number of example models to compare to: kilonovae, gamma-ray burst afterglows, shock cooling supernovae, core-collapse supernovae, etc.
We can demonstrate the functionality of the pipeline using a quick example. Taking the Metzger (2017) blue kilonova model as an example, we can generate a set of injections simply using the prior file (all are found in priors/).
nmma-create-injection --prior-file priors/Me2017.prior --eos-file example_files/eos/ALF2.dat --binary-type BNS --n-injection 100 --original-parameters --extension json
This generates a file called injection.json that includes an injection file drawn from the prior file with a number of injections specified by –n-injection.
It is this file that is used for the Bayesian inference analysis. An example analysis is as follows:
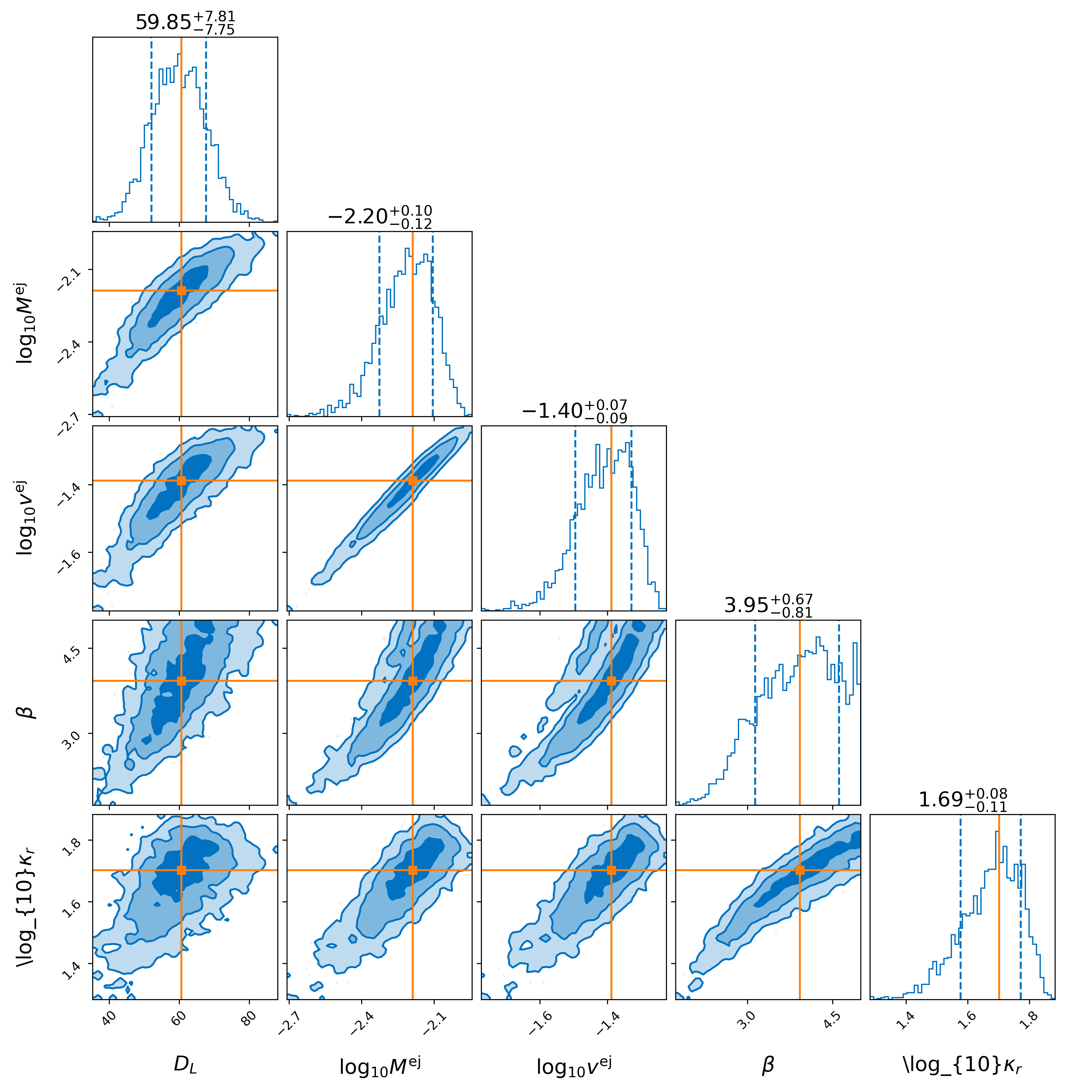
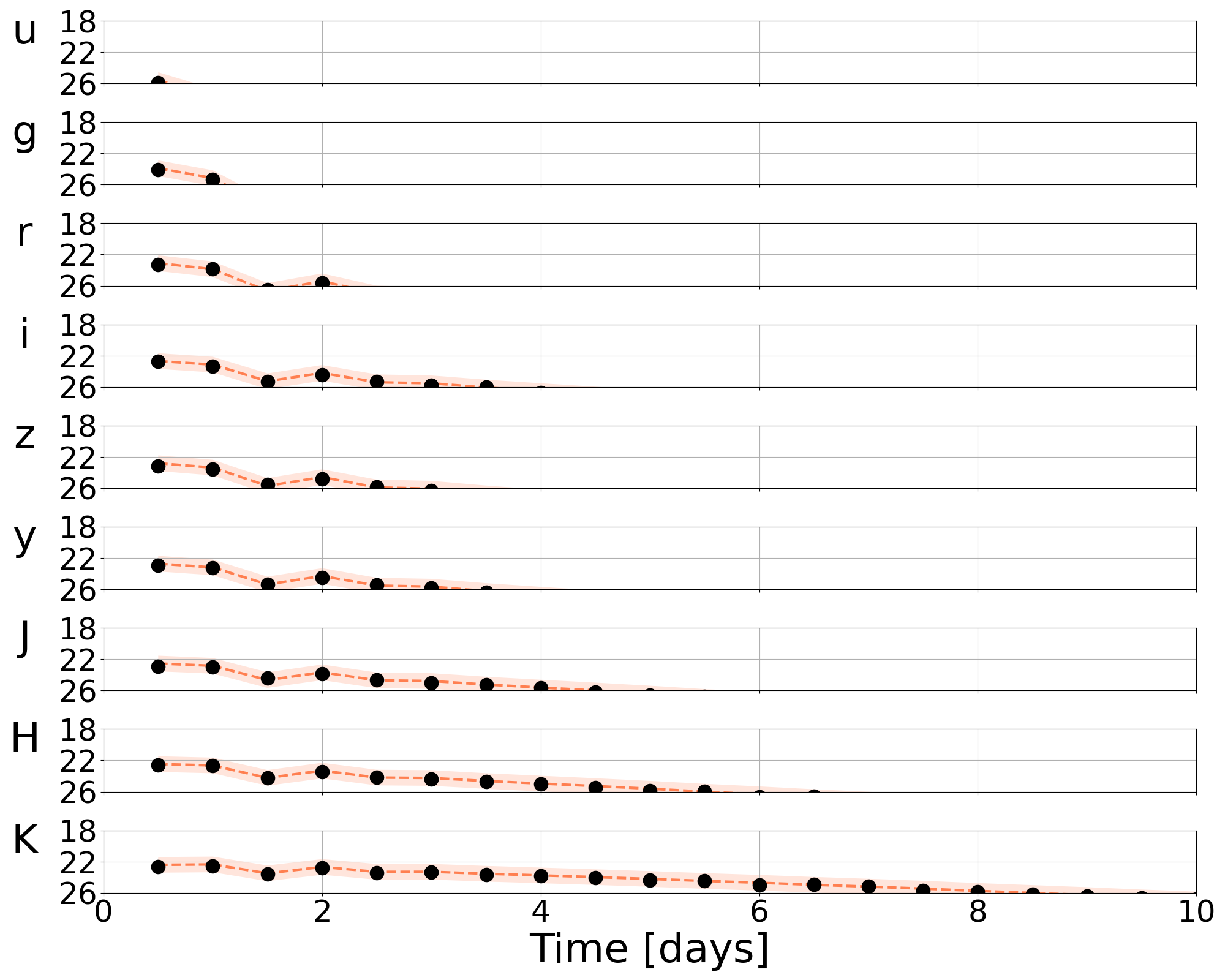
lightcurve-analysis --model Me2017 --outdir outdir --label injection --prior priors/Me2017.prior --tmin 0.1 --tmax 20 --dt 0.5 --error-budget 1 --nlive 512 --Ebv-max 0 --injection ./injection.json --injection-num 0 --injection-outfile outdir/lc.csv --generation-seed 42 --filters u,g,r,i,z,y,J,H,K --plot --remove-nondetections
Here, the time array is specified by a minimum, maximum, and delta t (in days) as specified by –tmin, –tmax, and –dt. The particular injection chosen is drawn from an index specified by –injection-num. The –filters available are specified with –filters u,g,r,i,z,y,J,H,K. Summary plots are available in outdir/.
Please note that Me2017 is a non-SVD model which means it does not need the –svd-path option to be specified. An SVD model example is as follows:
nmma-create-injection --prior-file priors/Bu2019lm.prior --eos-file example_files/eos/ALF2.dat --binary-type BNS --n-injection 100 --original-parameters --extension json --aligned-spin
lightcurve-analysis --model Bu2019lm --svd-path ./svdmodels --outdir outdir --label injection --prior priors/Bu2019lm.prior --tmin 0.1 --tmax 20 --dt 0.5 --error-budget 1 --nlive 512 --Ebv-max 0 --injection ./injection.json --injection-num 0 --injection-outfile outdir/lc.csv --generation-seed 42 --filters ztfg,ztfr,ztfi --plot --remove-nondetections --ztf-uncertainties --ztf-sampling --ztf-ToO 180